Research
Interests
My current research interests are primarily in developing tools in Topological Data Analysis (TDA) for computational biology. I am working with Guowei Wei on a couple of projects to build Persistent Topological Laplacians, like the one in this paper by Rui Wang.
Projects
Persistent Laplacian Library
I am working on a C++ library (with Python bindings) to efficiently compute many persistent Laplacian variations, unifying some disparate projects from multiple research groups in an efficient and user-friendly tool.
With preliminary testing, an experiment with the PDFL (below) that took 1.5 hours on 1 core of MSU’s High Performance Computing Cluster (HPCC) now takes about 12 seconds on 1 core of my laptop.
A Persistent Topological Laplacian for Directed Flag Complexes
Paper on arxiv, code on GitHub.
By combining persistent topological Laplacians with the directed flag complex (aka directed clique complex), we made the Persistent Directed Flag Laplacian (PDFL), which can glean information about the persistent homology and more. This builds on the fast persistent homology tool, Flagser, which was built similarly to Ripser: Computing Persistent Homology of Directed Flag Complexes by Daniel Lütgehetmann et al., 2020.
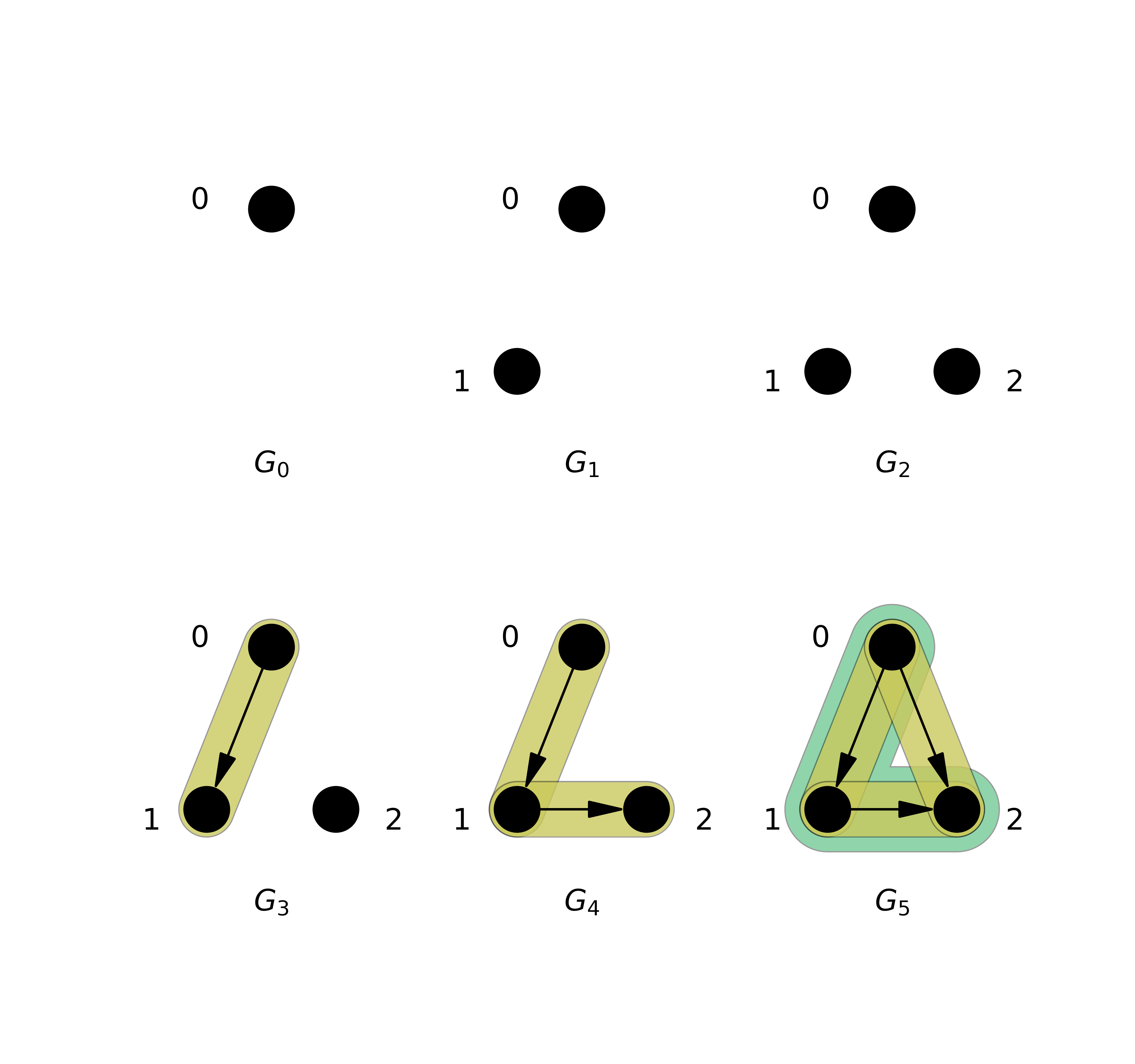
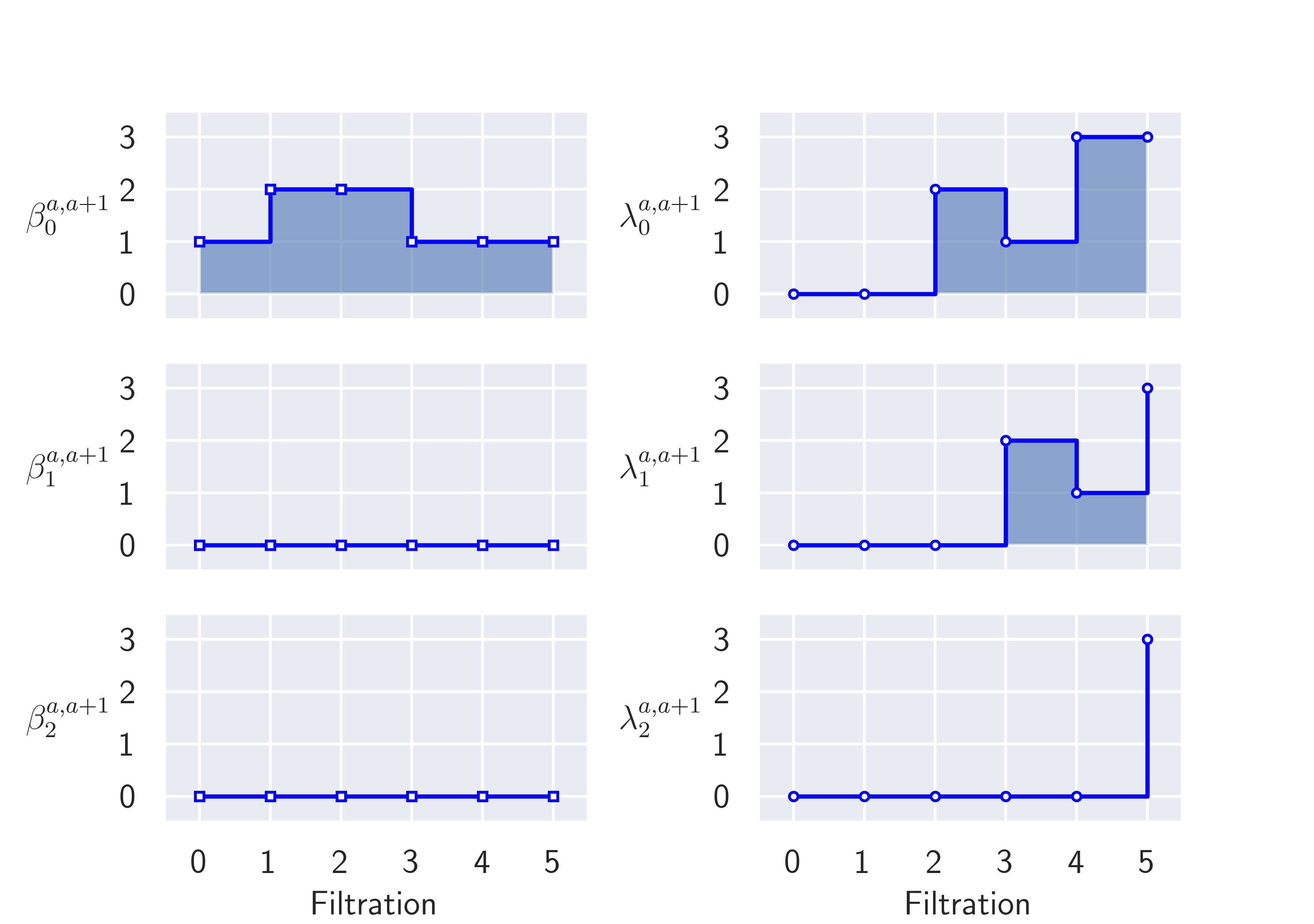
Here is a picture of a small filtered complex using a triangle, its persistent Betti numbers $\beta$ and the least nonzero persistent Eigenvalues $\lambda$.
Adaptive pseudo-time methods for solving the Nonlinear Poisson-Boltzmann Equation
This project is the basis for this paper, Ullah et al., 2023, and my master’s thesis. This work was completed with Shan Zhao’s research group at the University of Alabama.
I used a Proportional Integral Derivative (PID) controller to adaptively choose the time step in the numerical solution of a nonlinear PDE, the Poisson-Boltzmann Equation. This saved 84% of CPU time with only 0.06% relative error in computing the solvation energy of proteins.
